Master thesis
I did my master thesis under the supervision of Dr. Holger Fröhlich and Dr. Ashar Ahmad at the University of Bonn in Germany.
I presented the results of my master thesis at the following venues:
- 3rd Data Science Summer School at the Ecole Polytechnique - Paris, France - June 2019
- First Latin American Meeting on AI: KHIPU - Montevideo, Uruguary - November 2019
- AI Latin American Summit at the MIT Media Lab - Cambridge, USA - January 2020
Background:
The background of this master thesis is the Survival Based Bayesian Clustering (SBC) model developed by Ahmad and Fröhlich (2017), a model that infers clinically relevant cancer subtypes, by jointly clustering molecular data along with survival datain a semi-supervised manner. A graphical representation of the model is this:
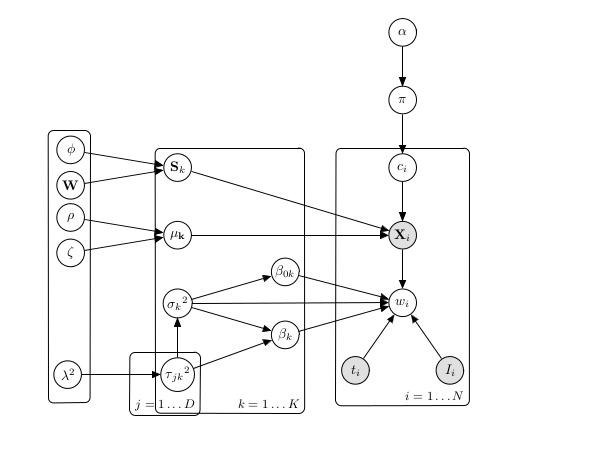
SBC’s main features are:
- Fully bayesian approach as omics data contains a lot of noise with p » n.
- Dirichlet Process prior to automatically infer the number of clusters.
- Molecular Data modelled as a Hierarchical Multivariate Gaussian Distribution (Mixture model).
- Survival time is modelled as Log-linear (Accelerated Failure Time) distribution with molecular covariates (Mixture model).
- L-1 regularization for the covariates of the Survival Model (Bayesian Lasso).
Abstract:
The SBC infers clinically relevant cancer subtypes, by jointly clustering molecular data along with survival data. Originally, the model was tested on a a Breast Cancer (Van De Vijver et al., 2002) and a Glioblastoma Multiforme (GBM) (Verhaak et al., 2010) data set, without any further external validation. The objective of this master thesis was to perform an external validation of the SBC, a goal that entailed two major tasks: a rigorous feature engineering and selection process that improved the known predictive ability of the model, and the characterisation of the obtained clusters and corresponding signature by delving into other types of clinical and omics data such as Copy Number Variation and miRNA.
The TCGA-GBM data set was retrieved using the Bioconductor package RTCGAToolbox and after data preprocessing, appropriate normalisation and correction for sample selection bias, a combined patient cohort of 421 samples was obtained (160 patients for the training and 261 patients for the validation set). Various feature engineering and selection techniques were explored. Every SBC model fit was done using Gibbs sampling. The best feature engineering and selection approaches were the Block HSIC-Lasso (Climente-González et al., 2019) model for mRNA-based selection and a Penalized Accelerated Failure Time (PAFT) model on a collection of oncogenic gene sets for pathway-based selection. In both cases there was an improvement of the initial Predictive C-Index (Block HSIC-Lasso feature selection = +1.5%, PAFT feature selection = +27.6%) and Recovery C-Index (Block HSIC-Lasso feature selection = +8.7%, PAFT feature selection = +5.0%).
The work done in this master thesis is a step forward in the validation of the SBC model on an external data set such as the TCGA-GBM patient cohort.
Written document:
If you would like to read my master thesis click here